Help
Code and data
For code and data including predictions of the human proteome please visit Zenodo at
Submit your sequences for prediction
You can submit one or multiple sequences (up to 100) by either copying and pasting them into the text box or uploading a file. In both cases, the sequences should be in FASTA format.
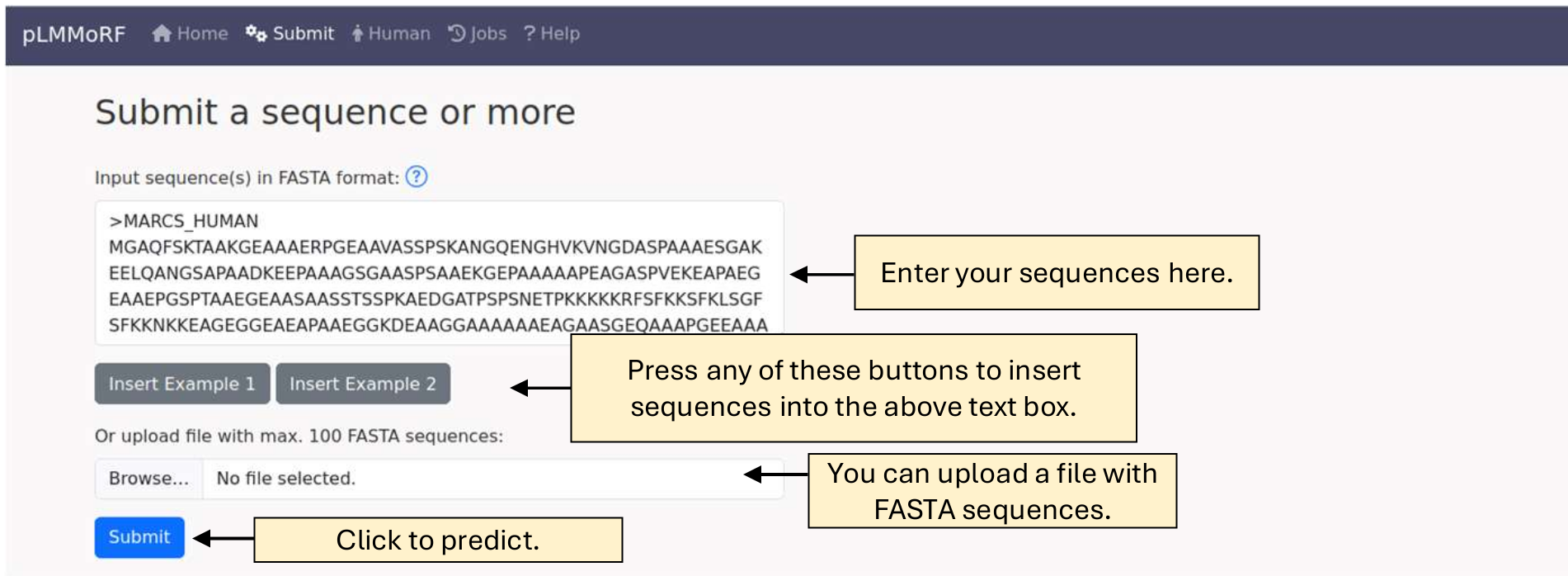
FASTA format overview:
(1) Each sequence begins with a header line, which starts with > followed by a unique ID (e.g., >Sequence1).
(2) The lines following the header contain the sequence in a single-letter amino acid code. The sequence can be split across multiple lines.
(3) Each new sequence should begin with its own header line.
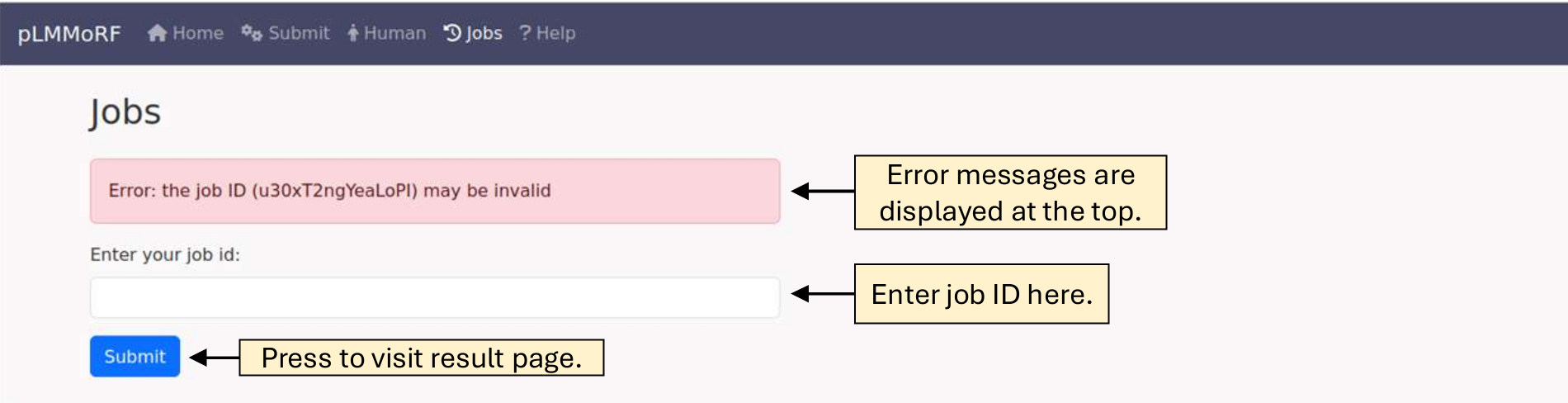
Check your results
If you have saved your Job ID, you can retrieve your results for up to 4 weeks by entering it into the text box on the Job page.
If you submitted multiple sequences, a table displaying your results will be provided.
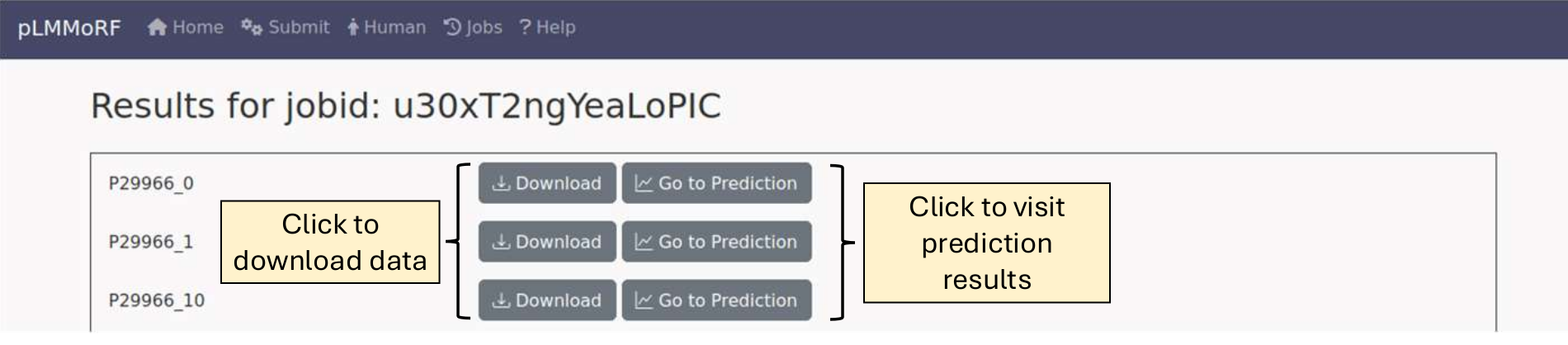
You can download individual results or all results, or view the predictions by selecting the appropriate button. An example prediction results page:
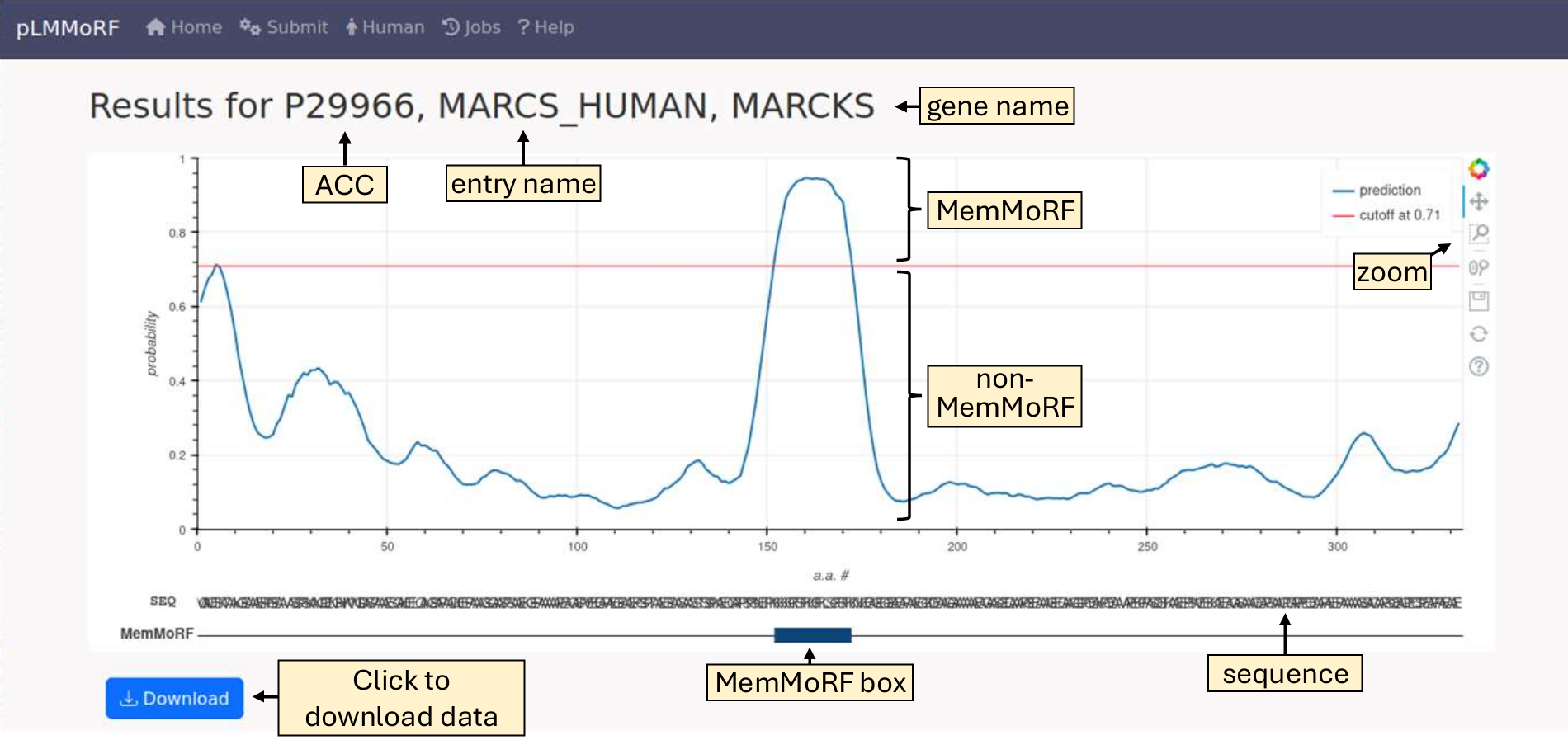
The MemMoRF box is red if the predicted region is shorter than 5 amino acids, indicating that shorter predictions are more likely to contain a higher number of false positives.
Look up human proteins
You can search for human proteins using various identifiers, including UniProt accession numbers (ACC), entry names, and gene names. The search is case-insensitive.
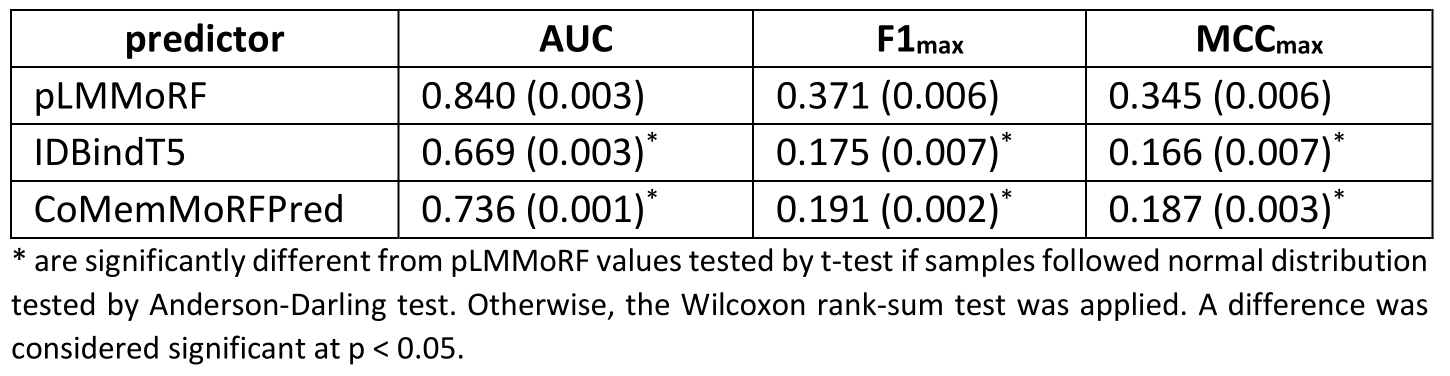
Performance and training/test sets
The datasets.zip can be downloaded from
containing json files and pickled pandas dataframes for:
(1) all proteins and residues,
(2) MemMoRF dataset of the pLMMoRF traing set,
(3) non-MemMoRF dataset of the pLMMoRF traing set, and
(4) test set.
The result JSON file
The dictionary loaded from the json file contains the following fields: (1) protein_id, (2) seq (the protein sequence), (3) proba (ordered list of probability values for each residue), and (4) pred (ordered list of binary predictions produced with a cutoff of 0.71).
Hardware
- Training was performed on an NVIDIA A100 GPU and took approximately four hours.
- Predictions are submitted and executed as follows:
- The FASTA sequence is written to a file within a directory named after the job ID. This directory is then copied to a dedicated server on the LAN.
- The job is launched via submission to SLURM, which is running on the LAN server equipped with an NVIDIA RTX 4000 SFF Ada GPU.
- The SLURM job submits the sequence to the pLMMoRF server program running on the same server. This server keeps the ProtT5 protein language model permanently loaded in GPU memory.
- The web application monitors a status file within the job directory on the LAN server.
- Once the job is marked as completed, the job directory's contents are copied back to the web server, and the result page is served.
Contact
hegedus.tamas@hegelab.org