Protein LM-based MemMoRF predictor
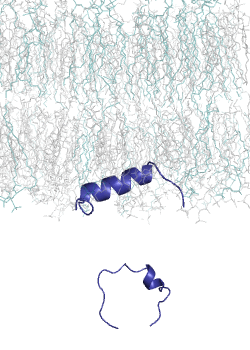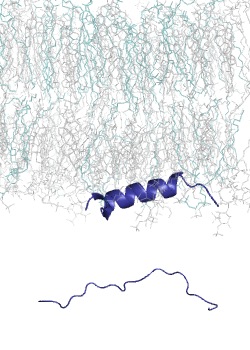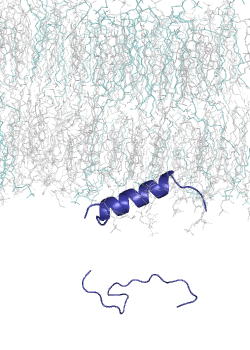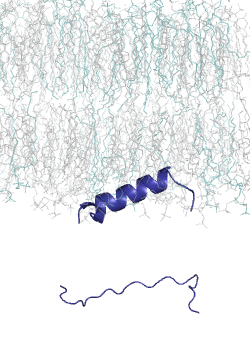
MemMoRF database is a curated database of membrane-associated proteins with regions that undergo structural changes when binding to membranes. These regions, called MemMoRFs (Membrane-binding Molecular Recognition Features), are critical in processes like signaling, membrane transport, and viral invasion. Many MemMoRF-containing proteins are linked to diseases through mutations, making them potential therapeutic targets. However, their flexible nature and membrane attachment pose challenges for drug discovery. The database compiles data from UniProt, PDB, and literature, focusing on proteins with NMR structures, along with disordered regions identified through X-ray and cryo-EM studies.
This set is small, thus we had developed a computational tool, CoMemMoRFPred to predict MemMoRFs. However, it is a meta-predictor with a large computational footprint. Therefore, we aimed to develop a more accurate and computationally efficient sequence-based predictor, utilizing embeddings from a protein language model (pLM) as input. Protein langaugae models built on vast amounts of protein sequence data, learning patterns and relationships across millions of sequences. By embedding sequences into high-dimensional spaces, pLMs capture subtle biological signals, such as disorder-to-order transitions, that are crucial for MemMoRF function.
- Predict your sequence: Upload your protein sequence to receive a prediction of potential MemMoRF regions.
- Look up human proteins: Search our precalculated database to see if your target protein contains any MemMoRF regions.
Publications:
-
J Mol Biol. 2025
[DOI: 10.1016/j.jmb.2025.169236]
[PMID: 40441416]
pLMMoRF: A web server that accurately predicts membrane-interacting molecular recognition features by employing a protein language model
M Csepi, B Berta, S Basu, L Kurgan, T Hegedus - J Mol Biol. 2023 Nov 1;435(21):168272.
[DOI: 10.1016/j.jmb.2023.168272]
[PMID: 37709009]
CoMemMoRFPred: Sequence-based Prediction of MemMoRFs by Combining Predictors of Intrinsic Disorder, MoRFs and Disordered Lipid-binding Regions.
Basu S, Hegedus T, Kurgan L -
Nucleic Acids Res. 2021 Jan 8;49(D1):D355-D360.
[DOI: 10.1093/nar/gkaa954]
[PMID: 33119751]
The MemMoRF database for recognizing disordered protein regions interacting with cellular membranes.
Csizmadia G, Erdos G, Tordai H, Padanyi R, Tosatto S, Dosztanyi Z, Hegedus T